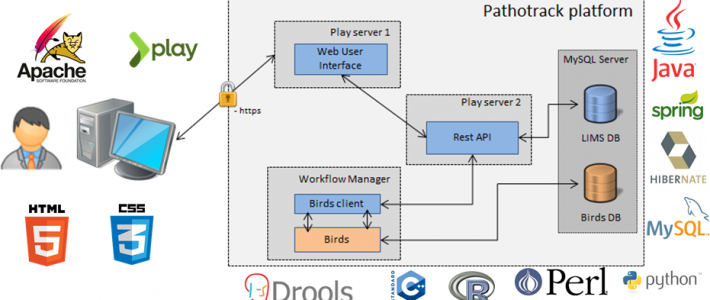
Ingénieur en développement logiciel pour la plateforme MicroScope
Localisé au CEA/Genoscope, un centre de recherche scientifique dédié à la génomique environnementale, le Laboratoire d’Analyses Bioinformatiques pour la Génomique et le Métabolisme (LABGeM, UMR8030, labgem.genoscope.cns.fr) s’intéresse à l’analyse et à l’exploration de la diversité des microorganismes et de leur