In addition to being time consuming and irrelevant for detection of a wide panel of pathogenic targets, current diagnostic methods for pathogens detection (microbial cultures, PCR-RFLP, test strips…) are almost useless in case of complex biological threats (e.g. several pathogens in a mixed sample). Thus, High-throughput sequencing (HTS) becomes undoubtedly a relevant tool for biodefense activities because besides being now affordable, it allows rapid identification of microorganisms without a priori, without isolation or prior cultivation, and can be even applied to microorganisms embedded in extremely complex matrices (i.e. metagenomic samples; e.g. soil samples, clinical samples, contaminated food…).
Some bioinformatics platforms/services dedicated to the detection of pathogens in metagenomic samples are beginning to emerge [1] but the robustness of the results obtained remains sometimes very controversial [2] due to either the quality of the sequences of the metagenomes analyzed or the choices of the comparison algorithms which often favor the speed to the precision. Despite these pitfalls, many service offers providing complete analysis of metagenomic samples have appeared on the web. Though practical, these solutions have however a number of disadvantages such as billing per batch samples or the impossibility of carrying out tailored analyzes. Furthermore, outsourcing such a service may also raise problems of data confidentiality.
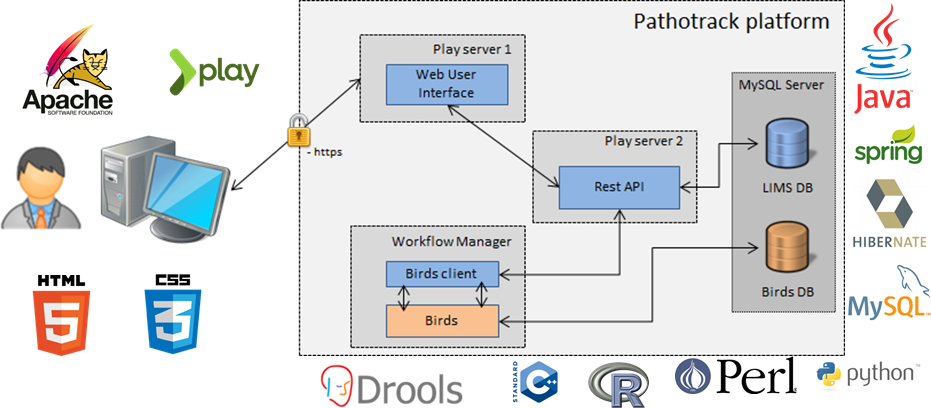
The combination of NGS analyses with cutting edge bioinformatics is at the heart of the PathoTRACK project which purpose is to develop a standalone (which does not require any special knowledge in bioinformatics and can be set up easily), scalable (to realize on-demand services) and portable (for in situ analyses) software/hardware solution allowing the rapid identification of threat agents in complex metagenomic samples. Our strategy is to take advantage of recent technology improvements, in term of sequencing (for example the MinION from Oxford Nanopores Technologies®) and bioinformatic tools, for biodefense objective but as well as for diagnostic prevention and treatment objectives.
References
1- (Naccache et al., 2014, 24 (7): 1180-92)
2- (Breitwieser et al., 2015, f1000research.6743.2)
