The proportion of protein sequences of unknown function in public databases stills very
important (42% of UniProt sequences are labelled as “hypothetical”, “uncharacterized”, “unknown” or
“putative”). On the other hand, a number of enzyme activities (about 30%) remain orphan (i.e. there is
any known sequence that is linked to this activity). Conserved functional modules identification in the
metabolism is one of the possible ways to improve protein functional annotation, by discovering new
enzyme reactions and new metabolic pathways. It is in this context that has been developed my PhD
thesis, proposing a new representation of the global metabolic network, where reactions sharing the same
chemical transformation type are grouped in reaction molecular signatures (RMS). A reaction signature is
the difference of its products and substrates stereo signatures molecular descriptors involved in this
reaction (Carbonell et al. 2013, http://molsig.sourceforge.net). These RMS are computed for all well
balanced reactions involved in at least one metabolic pathway, for which all substrates and products are
identified and have an available structure. RMS allow reaction classification in an automatic and expert-
independent way and a greater coverage of all enzymatic reactions that the classification of the Enzyme
Commission (EC numbers).
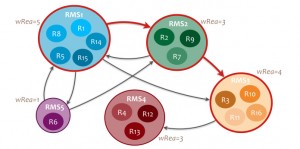
node, and edges conserve the initial connectivity between reactions. Several scores are then computed for
each path in the RMS network in order to assess known metabolic pathways conservation and to
discover new ones. The first score, scoreRea, is computed using the average reaction number by RMS
and represents the chemical conservation of the path in the whole metabolism. The second one,
scoreProt, is based on the protein number associated to each RMS and reflects the enzyme conservation
of the path through the tree of life. The next score, scoreTopo, is based on the PageRank centrality and
depicts the topological importance of an RMS sequence in the metabolic network. The last metric, the
Pathway Conservation Index (PCI) is the number of different reaction paths among known metabolic
pathways grouped in a same RMS path. It represents the conservation of chemical transformation
sequences in the known part of the metabolism. Most conserved RMS paths are next identified in order
to understand the linkage between different conservation types (chemical, enzymatic and topologic) and
the biological processes type of metabolic pathways (like biosynthesis or degradation).
This metabolism representation has an interesting predictive potential and can be used to identify most
conserved parts of the metabolism and to discover new metabolic modules. Moreover, combination of
different scores can be used to predict the metabolic role of new pathways using machine learning
approaches. Conserved paths of chemical transformations associated to genomic context data will be a
useful tool for functional annotation of genes and groups of genes of unknown function.