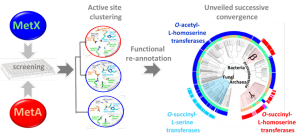
MetX and MetA are two phylogenetically unrelated protein families, but both are implied in the first step of the L-methionine biosynthesis pathway. MetX proteins are known as L-homoserine O-acetyltransferases (HAT), while MetA proteins are generally annotated as L-homoserine O-succinyltransferase (HST). Here we demonstrated that in vitro, over ~100 enzymes, that HAT and HST activities are actually widely observed in both families. In silico analysis of active site structures pointed out key residues specific to each activity. Thus, we reexamined nearly 10 000 MetA and MetX proteins using homology modeling and we corrected the function for about 60% of them. The mapping of these predictions on the NCBI taxonomy tree shows that ancestral MetX and MetA had both the same activity, and suggests that an evolutionary pressure led them to evolve toward HTS activity in proteobacteria. We finally discovered that 10% of MetX are paralogs that participate in L-cysteine synthesis in fungi and probably in few gammaproteobacteria by forming the novel metabolite O-succinyl-L-serine.
People: Karine Bastard, Alain Perret et Véronique de Berardinis (UMR 8030 Métabolique Génomique du Genoscope)
Publication: Bastard K, Perret A, Mariage A, Bessonnet T, Pinet-Turpault A, Petit JL, Darii E, Bazire P, Vergne-Vaxelaire C, Brewee C, Debard A, Pellouin V, Besnard-Gonnet M, Artiguenave F, Médigue C, Vallenet D, Danchin A, Zaparucha A, Weissenbach J, Salanoubat M, de Berardinis V. Parallel evolution of non-homologous isofunctional enzymes in methionine biosynthesis. Nat Chem Biol. 2017 Jun 5. doi: 10.1038/nchembio.2397.
Website : http://www.genoscope.cns.fr/metgenes/