Annotation and analysis of microbial genomes are the core of our scientific activities, especially through collaborative projects using the MicroScope platform. They often involve comparative and phylogenetic analysis to understand dynamics and evolution of bacterial genomes, syntactic and functional annotation of new bacterial genomes or genomes from reference organisms (i.e., E. coli, B. subtilis, P. putida). To face the challenge of the analysis and visualisation of thousands microbial genomes, new methodological developments are addressed.

Multi-enzymatic nanomachines for the controlled transformation of terrestrialplant biomass.
The climate emergency requires us to move from an economy based on fossil fuels to a sustainableenergy mix, allowing a transition from our carbon-based societies to a more sustainable economy.In this context, the valorisation of lignocellulosic biomass (LCB) is central for the development ofthe circular bioeconomy. LCB is composed of ...
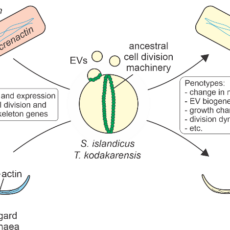
ARCHANE : Reconstructing the cell biology of the archaeal ancestor of eukaryotes
Abstract The emergence of eukaryotes (eukaryogenesis) is one of the central, extensively debated and yet unanswered questions in biology. It was proposed that informational genes as well as cytoskeleton and membrane remodelling/cell division machineries of eukaryotes have been inherited from an archaeal ancestor. Most of these eukaryotic-like proteins, often referred ...

MUDIS4LS: Mutualised Digital Spaces for FAIR Data in Life and Health Sciences
MUDIS4LS project (ESR+/EquipEx PIA3) is leaded by the French Institute of Bioinformatics (IFB). It is based on 4 technological workpackages and 5 thematic projects (Implementation Studies). It gathers 39 teams from 14 organisations, 4 national data centers, 7 regional data centers and 6 national data-producing infrastructures. In addition to the ...

ABRomics: the national multiomics platform for Antibiotic Resistance research and surveillance
Funded by the French Priority Plan on antibioresistance, the ABRomics project aims at developing an online community-driven platform to scale up and improve surveillance and research on antibiotic resistance from a One Health perspective. The platform is made of: A repository of structured, interoperable, standardized and well-annotated multi-omics microbiological data ...

BlueRemediomics: Harnessing the Marine Microbiome for Novel Sustainable Biogenics and Ecosystem Services
Launched in December 2022, the Horizon Europe BlueRemediomics project aims to harness the untapped potential of marine microbial resources. Lasting four years, and bringing together an international consortium of experts including the Metabolic Genomics UMR (Genoscope/CEA-Jacob), this project will develop new tools and new approaches to explore marine microbiome data ...
CarboMagnet : Contribution of magnetotactic bacteria forming intracellular carbonate mineral phases to the sequestration of organic carbon and alkaline earth elements
Abstract The project focuses on a poorly known carbon-sequestering environmental process by a group of diverse bacteria forming intracellular carbonates, occurring at oxic/anoxic boundaries (OAB) in aquatic environments. The molecular and geochemical processes of bacterial biomineralization will be studied by a combination of approaches in isotope and aqueous geochemistry, mineralogy, ...
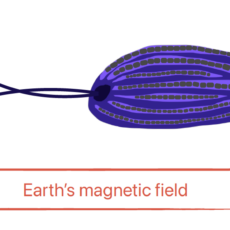
SymbioMAGNET : Deciphering the biodiversity, ecology and evolution of magnetotactic symbiosis
Abstract The project aims to decipher the biodiversity, ecology and evolution of a group of unicellular eukaryotes ubiquitous in marine anoxic sediments that sense the Earth’s magnetic field thanks to their symbiotic bacteria. This exploration will be done with a combination of approaches in microbiology, microscopy and genomics from the ...
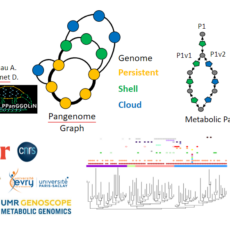
ModelOmics
Background Today, thanks to the development of metagenomics, our knowledge of microbial diversity has greatly changed. Whereas previously cellular organisms were seen more as independent entities, today we know that they organize themselves into complex microbial communities made up of several species of bacteria, archaea and eukaryotes, and their mobilome ...
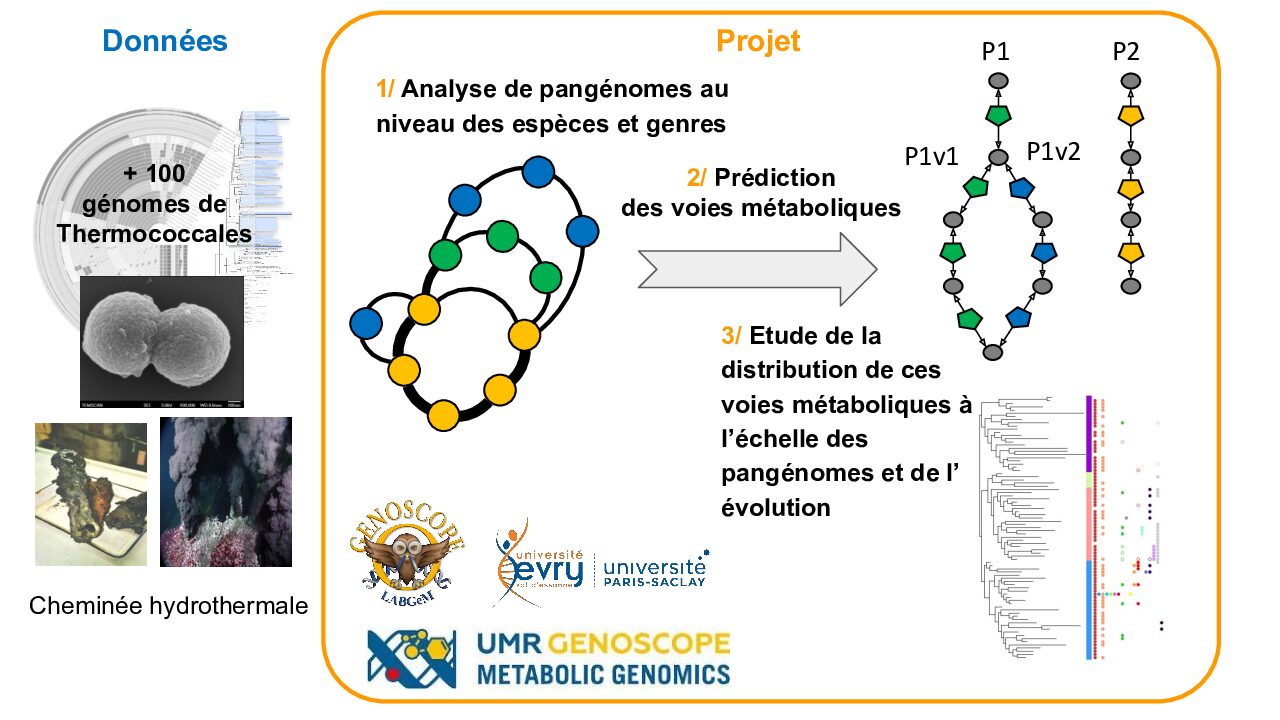
PanGenoThermo – Pangénomique et Métabolisme chez les Thermococcales – Clémence Lauden
Description Contexte scientifique : Les Thermococcales sont un ordre d’archées anaérobies et hyperthermophiles, qui vivent au niveau des cheminées océaniques hydrothermales et au niveau de sources chaudes terrestres à pH neutre 1–3. Elles sont utilisées comme organisme modèle pour l’étude de la vie à haute température (avec une température optimale ...
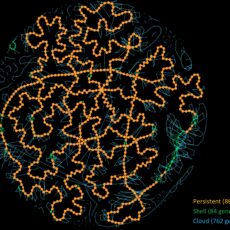
PanGBank: a comprehensive database for pangenome analysis and exploration
Many comparative genomic studies try to get a grasp on the overall gene content of a species. However, with the current explosion of available genomic data, it becomes more complex to use all-vs-all genome comparison approaches. Over the last years, the concept of pangenome emerged, whose goal is to capture ...
