Genomics
Next-generation sequencing (NGS) technology has revolutionized genomics leading to an unrivaled generation of large overloads of omics data ((meta)genomes, (meta)transcriptomes, epigenomes). The use of sequencing machines, which are now smaller in size but capable of generating piles of data faster and at a lower cost, have however raised the challenges of sharing, archiving, integrating and analyzing these data. Efficient and effective pipelines/tools to deal with those increasing amounts of genomics data are therefore needed to store, transfer, analyze, visualize, and generate reports to researchers and possibly clinicians. In this scope, at LABGeM we develop standard as well as innovative tools to get the most of NGS data.

MUDIS4LS: Mutualised Digital Spaces for FAIR Data in Life and Health Sciences

ABRomics: the national multiomics platform for Antibiotic Resistance research and surveillance
The platform is made of:
- ...
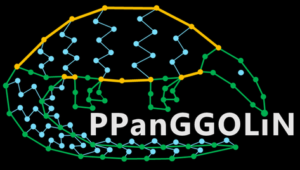
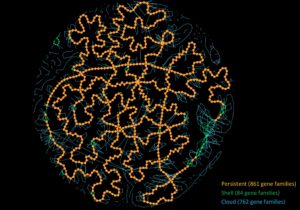
PPanGGOLiN: Depicting microbial species diversity via a Partitioned PanGenome Graph Of Linked Neighbors
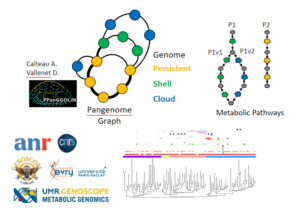
ModelOmics
PanGBank: exploring GenBank genomes through a resource of pangenome graphs
Functional Annotation
High quality functional annotation is essential for understanding the phenotypic consequences encoded in a genome. We develop several methods to improve protein function predictions. These methods are mainly based on sequence analysis, genomic and metabolic context exploration and structural bioinformatics.
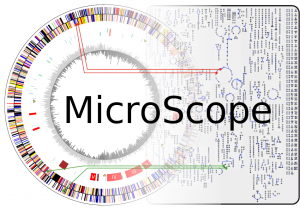
Prediction, annotation and exploration of genomic regions in MicroScope

