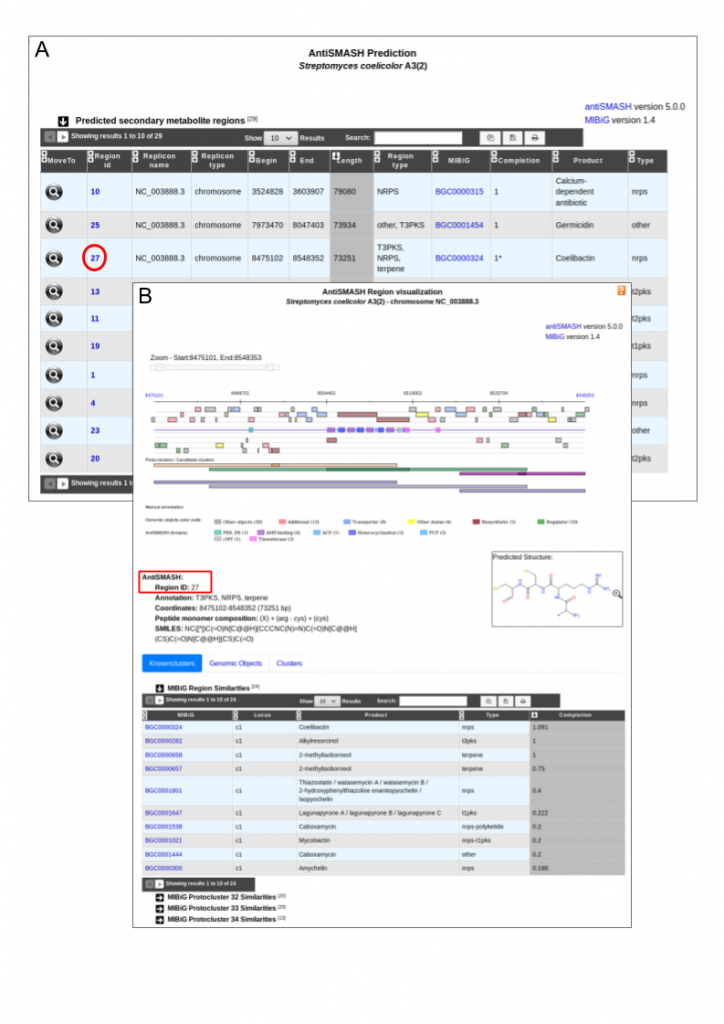
The secondary metabolite gene cluster prediction tool antiSMASH has been updated to version 5 on the MicroScope platform. This new version includes updated and extended cluster identification profiles, and allows to detect 52 types of Biosynthetic Gene Clusters (BGCs) [1].
The program also offers extended and improved BGC detection and analysis capabilities particularly for regions containing multiple BGCs of one or several types. To help users analyze these regions, the program introduces a new terminology to describe different biological options that lead to BGCs with notably ‘protoclusters’ and ‘candidate clusters’. Hence, the MicroScope graphical interface for the visualization of predicted secondary metabolite clusters has been updated to show these (see figure below). This new feature is particularly useful for the analysis of complex regions composed of multiple BGCs.
Moreover, corresponding result tables have been split into three sections to describe:
- similarities with known clusters from the MIBiG database,
- the list of genomic objects that compose the region and
- candidate cluster and protocluster coordinates.
Reference:
[1] Blin K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019 Jul 2;47(W1):W81-W87.
