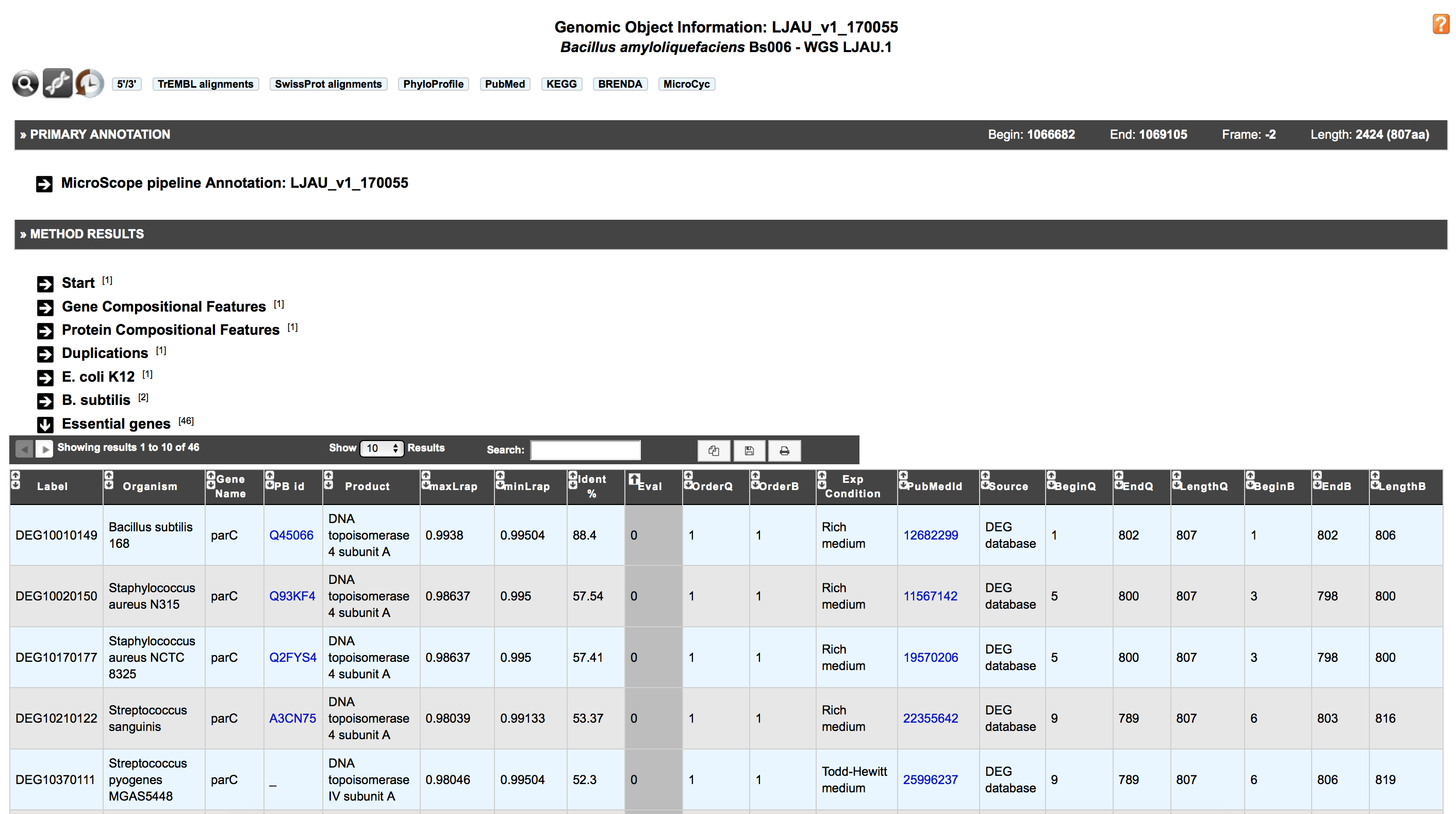
Delineating a set of essential genomic elements and proteins that make up a living organism helps to understand critical cellular processes that sustain life. Essential genes in a bacterium constitute a minimal genome, forming a set of functional modules, which play key roles in the emerging field of synthetic biology. A new ‘essential gene’ dataset coming from Database of Essential Genes (DEG) has been set up in MicroScope. This dataset comes in replacement of the 3 datasets from the 3 model organisms P. aeruginosa PAO1, E. coli K12 and B. subtilis 168.
The DEG hosts records for bacterial and archaeal essential genes determined by genome-wide gene essentiality screens. These essential genomic elements have been determined not only in vitro, but also in vivo, under diverse conditions including those for survival, pathogenesis and antibiotic resistance.
Moreover the DEG dataset has been improved with data from Acinetobacter baylyi ADP1 and Neisseria meningitidis 8013, two highly curated genome in MicroScope.
‘Essential genes’ results are available for each CDS of all genomes and they can also be queried through the ‘Search by keywords‘ functionality.
References:
Hao Luo, Yan Lin, Feng Gao, Chun-Ting Zhang and Ren Zhang, (2014) DEG 10, an update of the Database of Essential Genes that includes both protein-coding genes and non-coding genomic elements. Nucleic Acids Research 42, D574-D580.
de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, Le Fèvre F, Schächter V, Pezo V, Döring V, Scarpelli C, Médigue C, Cohen GN, Marlière P, Salanoubat M, Weissenbach J. (2008) A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol.;4:174.
Rusniok C, Vallenet D, Floquet S, Ewles H, Mouzé-Soulama C, Brown D, Lajus A, Buchrieser C, Médigue C, Glaser P, Pelicic V. (2009) NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol.;10(10):R110.
