 AntiSMASH (antibiotics & Secondary Metabolite Analysis Shell) [1,2] is now available on MicroScope. This program allows the rapid genome-wide identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genomes. It integrates (and cross-links with) a large number of in silico secondary metabolite analysis tools that have been published earlier.
AntiSMASH (antibiotics & Secondary Metabolite Analysis Shell) [1,2] is now available on MicroScope. This program allows the rapid genome-wide identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genomes. It integrates (and cross-links with) a large number of in silico secondary metabolite analysis tools that have been published earlier.
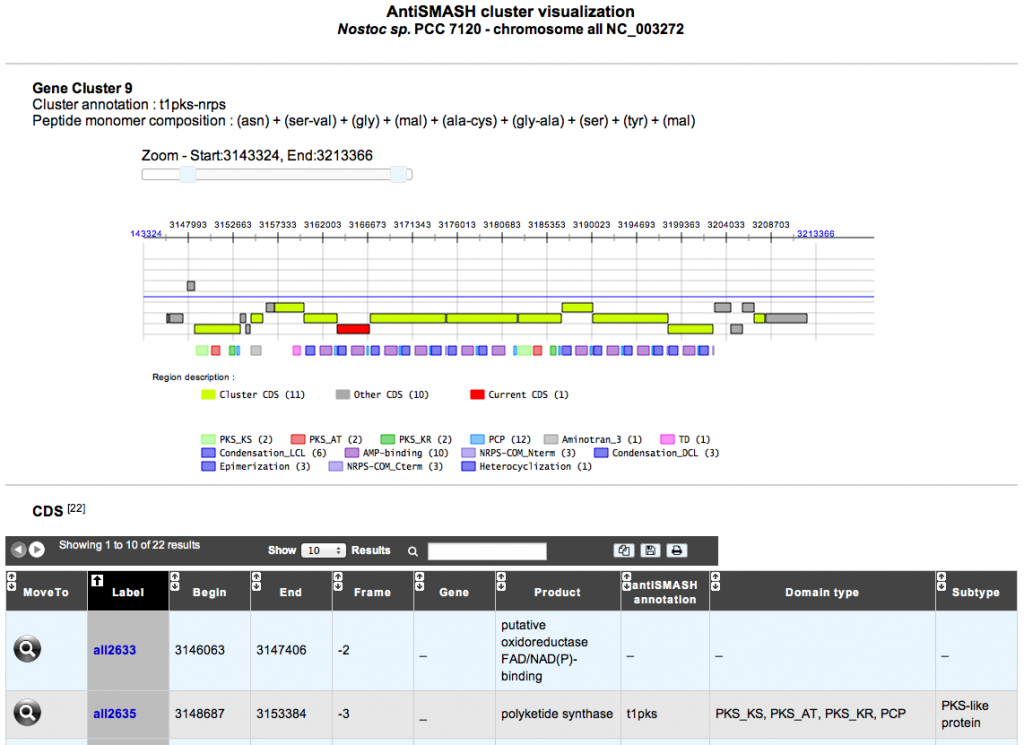
AntiSMASH detects biosynthetic loci covering the whole range of known secondary metabolite compound classes. Its results are available in the gene annotation window and can also be queried using the ‘Search by keyword’ functionality. You also have the possibility to visualize each antiSMASH cluster prediction and its genomic context. In case of NRPS/PKS cluster type, the predicted peptide monomer composition is indicated as well.
[1] Blin K., et al. (2013) antiSMASH 2.0 — a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Research. Jul;41(Web Server issue):W204-12
[2] Medema M.H., et al. (2011) antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters.Nucleic Acids Research. Jul;39(Web Server issue):W339-46.
