Retirement of MICHECK and AMIGene
Dear users, We have decided to retire our old web tools MICHECK and AMIGene. Those tools have not been maintained for several years and are no longer useful as they (and many others) are integrated in MicroScope. We will proceed ...
PPanGGOLiN: Depicting microbial species diversity via a Partitioned PanGenome Graph Of Linked Neighbors
PPanGGOLiN (Gautreau et al. 2020) is a software suite used to create and manipulate prokaryotic pangenomes from a set of either genomic DNA sequences or provided genome annotations. It is designed to scale up to tens of thousands of genomes ...
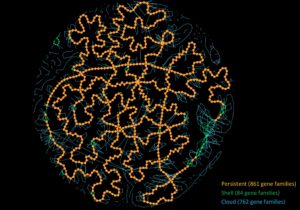
PanGBank: a comprehensive database for pangenome analysis and exploration
Many comparative genomic studies try to get a grasp on the overall gene content of a species. However, with the current explosion of available genomic data, it becomes more complex to use all-vs-all genome comparison approaches. Over the last years, ...

ASMC – Classifying Active Sites of Enzymes Families
ASMC method (Active Sites Modeling and Clustering) is a novel unsupervised method to classify sequences using structural information of protein pockets. The method predicts functional amino-acids by proposing active site SDP residues (Specificity Determining Position) and active site CP residues ...
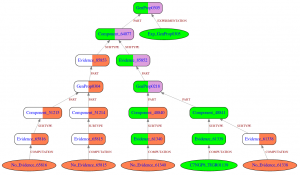
GROOLS: Reactive Graph Reasoning for Genome Annotation
GROOLS (Genomic Rule Oriented Object Logic System) is a bioinformatics software that helps biologists in the evaluation of genome functional annotation through biological processes like metabolic pathways. GROOLS is an expert system that uses paraconsistent logic. It evaluates the completeness ...

NetSyn – Exploring Syntenies Networks of Genes
We developed a method to classify proteins of a family based on their conserved genomic contexts. Each protein genomic context is compared against all others to determine syntenies. A graph is generated where nodes are input proteins connected by edges ...
