From annotation and analysis of microbial genomes, Genome-Scale Metabolic Networks (GSMN) can be obtained and contain all of the known metabolic reactions in a given organism and the genes that encode each enzyme. Part of our research activities are focused on the curation of GSMN and the development of databases and methods to improve their reconstruction.
Two main bottlenecks limit the understanding of metabolic consequences encoded in a genome: the difficulty in associating correct functions to genes and the lack of experimental characterization of enzyme activities for which proteins are sometimes unknown, i.e. orphan enzymes. In the light of enzymatic screening and metabolomics experiments, we develop bioinformatics methods and perform bioanalysis for the discovery of new enzyme families and the exploration of their functional diversity. Methods are mainly based on sequence analysis, genomic and metabolic context exploration and structural bioinformatics.

The climate emergency requires us to move from an economy based on fossil fuels to a sustainableenergy mix, allowing a transition from our carbon-based societies to a more sustainable economy.In this context, the valorisation of lignocellulosic biomass (LCB) is central for the development ofthe circular bioeconomy. LCB is composed of ...

Launched in December 2022, the Horizon Europe BlueRemediomics project aims to harness the untapped potential of marine microbial resources. Lasting four years, and bringing together an international consortium of experts including the Metabolic Genomics UMR (Genoscope/CEA-Jacob), this project will develop new tools and new approaches to explore marine microbiome data ...
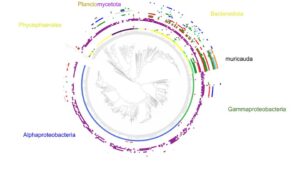
The Phaeoexplorer project (https://phaeoexplorer.sb-roscoff.fr/home/) aims to generate transcriptome data and annotated genome assemblies for a broad range of brown algal species at different phylogenetic distances from the model brown alga Ectocarpus. It is made in collaboration with the algal genetics group in Roscoff (https://www.sb-roscoff.fr/en/team-algal-genetics). During the sequencing step, we also ...
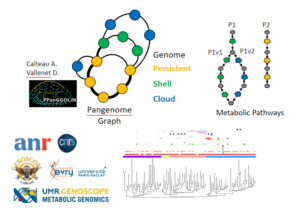
Background Today, thanks to the development of metagenomics, our knowledge of microbial diversity has greatly changed. Whereas previously cellular organisms were seen more as independent entities, today we know that they organize themselves into complex microbial communities made up of several species of bacteria, archaea and eukaryotes, and their mobilome ...
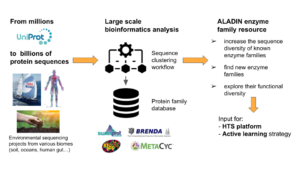
The ALADIN project aims to build an integrated and distributed technological platform which will rely on the combination of combinatory genetics with artificial intelligence. It will enable the exploration of natural molecular diversity, its appropriation to build new purpose-based catalysts (enzymes and micro-organisms), and the rapid expansion of the knowledge ...
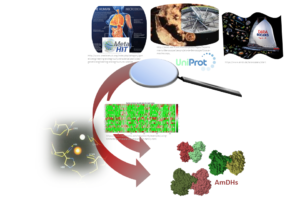
The current trend in Chemistry is to perform the energy transition successfully by considering cheaper and greener alternatives. This can be achieved by the substitution of some chemical steps by biocatalyzed ones or the design of new enzymatic routes, thus reducing waste and polluting organometallic catalysts. As useful building blocks ...

We developed a method to classify proteins of a family based on their conserved genomic contexts. Each protein genomic context is compared against all others to determine syntenies. A graph is generated where nodes are input proteins connected by edges when a synteny is observed. Edges weight represent the mean ...







