The secondary metabolite gene cluster prediction tool antiSMASH has been updated to version 4 on the MicroScope platform. This new version includes updated and extended cluster identification profiles, especially for ribosomally synthesised peptide products (RiPPs) [1]. The ClusterFinder algorithm is also used to try to improve prediction of gene cluster boundaries. These estimations are based on frequencies of locally encoded protein domains detected by Pfam (based on these being either more or less BGC-like).
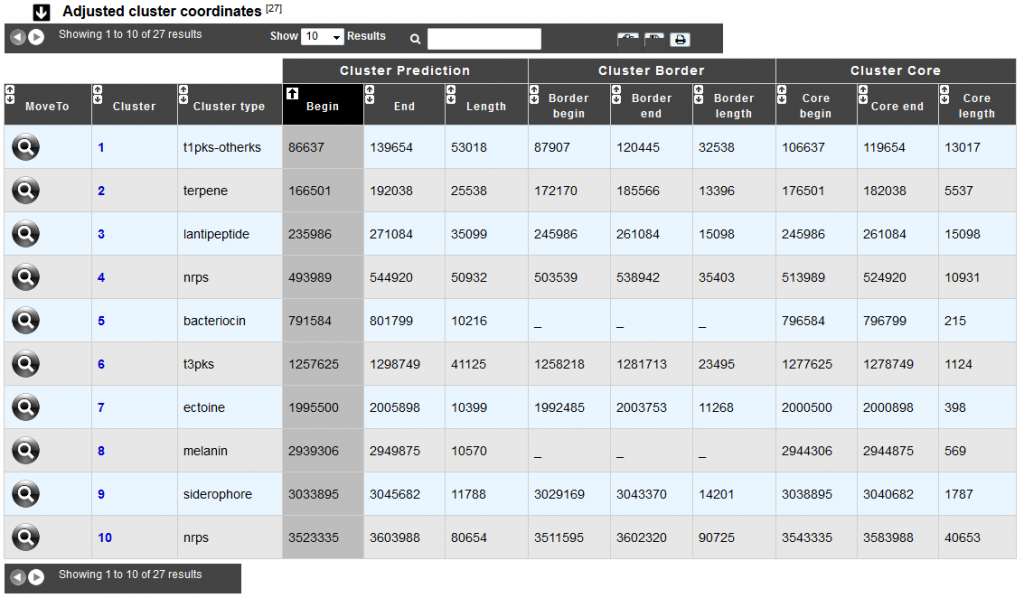
The alternate coordinates are indicated in the “Adjusted cluster coordinates” table as well in the cluster visualization window.
Moreover predicted clusters are compared to a more recent version of the reference database MIBiG (Minimal Information about a Biosynthetic Gene cluster), which describe more than 1400 biosynthetic gene clusters (BGC) of all types (NRPS, PKS or RIPP) and from all phyla (bacteria, fungi or plants).
References:
[1] Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017 Jul 3;45(W1):W36-W41.