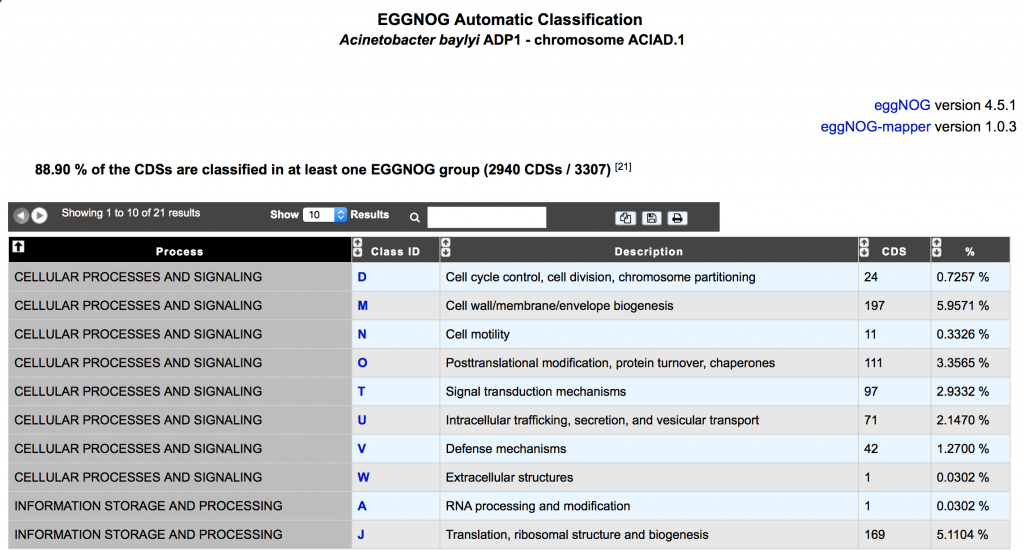
eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) is a public resource that provides Orthologous Groups (OGs) of proteins at different taxonomic levels, each with integrated and summarized functional annotations.
eggNOG uses a graph-based unsupervised clustering algorithm extending the COG methodology to produce genome wide orthology inferences, which are further adjusted to provide lineage specific resolution. The database currently covers 2031 eukaryotic and prokaryotic organisms, as well as precomputed mappings for 1655 additional prokaryotes
eggNOG version 4.5.1 is now available on the platform allowing new functional annotations and classification.
More: http://eggnogdb.embl.de/#/app/methods
Reference: Huerta-Cepas J, et al. eggNOG 4.5: a hierarchical orthology framework with improved functionalannotations for eukaryotic, prokaryotic and viral sequences. Nucl. Acids Res. (04 January 2016) 44 (D1): D286-D293.