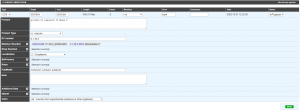
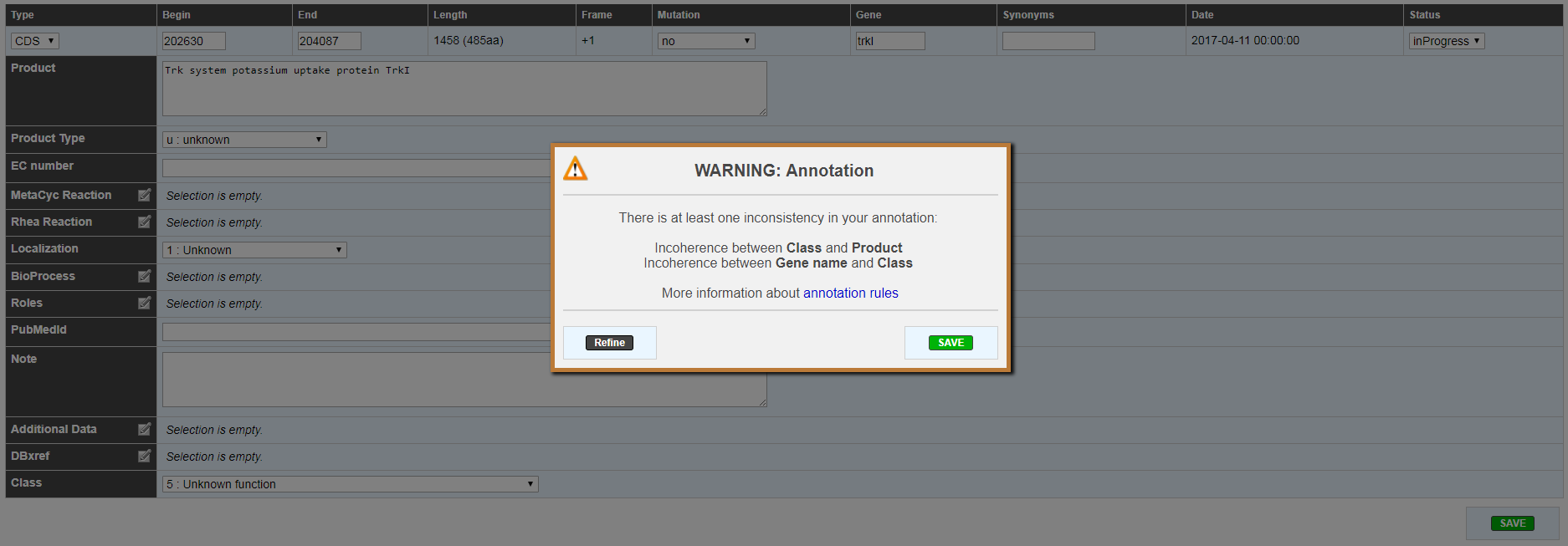
The gene annotation editor has been improved with a more accurate form and a better inconsistency check.
The form will adapt to the object type you are annotating. For instance, you won’t have the same editable fields if you are annotating a CDS or a tRNA.
The “note” field is now editable, it corresponds to the “/note” qualifier in genbank/EMBL format and therefore it will be exported into submission files (coming soon).
The “comment” field has been replaced by “additional data”. This new section allows to store multiple types of data such as : comments, interaction data, phenotype data or SMILES formulas.
The new “dbxref” field allow you to link external database elements related to your annotation. In the future it will be exported into submission files in /dbxref qualifier of genbank/EMBL.
In the “Product type” list, the putative elements have been deleted. In order to annotate putative element, you have to write ‘putative protein of …’ in the “Product” field while having “class” set at ‘3: Putative function from multiple computational evidences’. You can access to all annotation rules here.
Annotation inconsistency check has been improved in order to warn you before saving and give you field name that are in conflict.
You can propose new elements to add in “additional data” and “dbxref” fields here