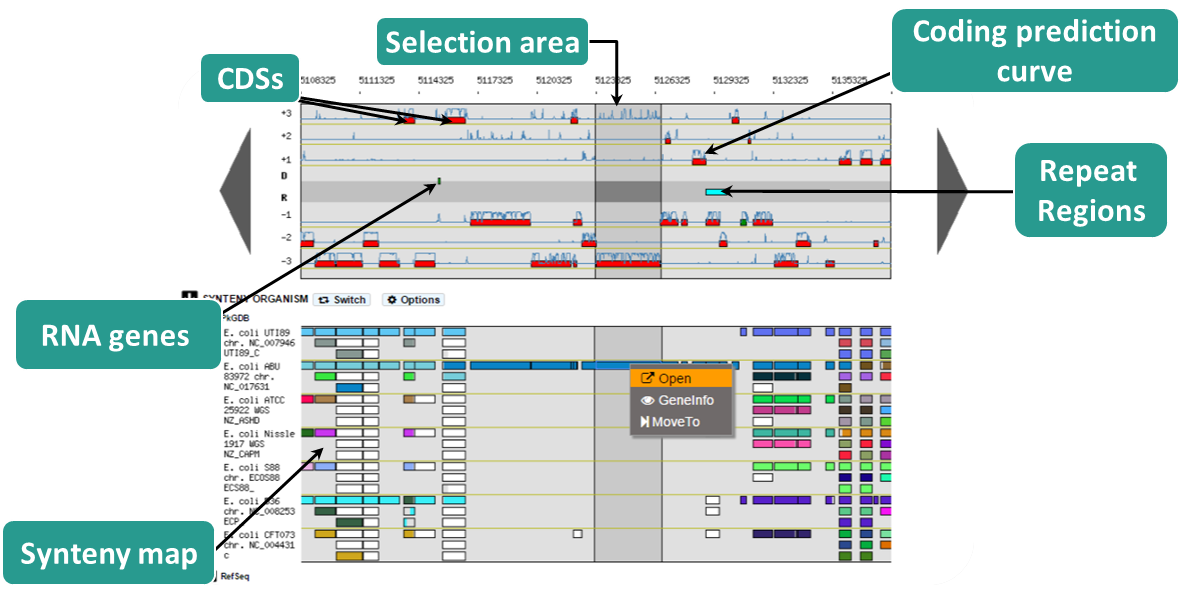
The Genome Browser has been updated to enhance visibility of genomic objects. In the viewer you can notice that the central part is now grey to separate the reverse strand from the direct one. In this central part are represented repeat regions as well as genomic object (tRNA, rRNA, misc_RNA, tmRNA, ncRNA, IS, misc_feature, promoter) which use strand (+1/-1) attribute instead of frame.
New functionalities have also been added to ease the navigation and exploration of genomic objects in the genome browser and in the pkgdb synteny maps.
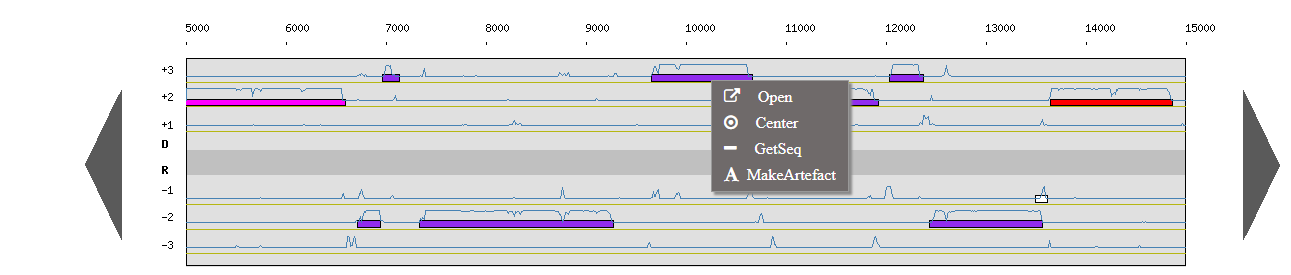
A contextual menu has been added to these two windows, which mean that you can right click on a genomic object to navigate. Here is the list of available options :
- New: create a new object (on genome browser background ; with annotation rights only)
- Open: open the gene/synteny information page (on object in both windows)
- Center: center and highlight the object (on genome browser object)
- GetSeq: obtain the object sequence (on genome browser object)
- MakeArtefact: transform the object into Artefact (on genome browser object ; with annotation rights only)
- GeneInfo: open the gene information page (on pkgdb synteny object)
- MoveTo: go to the selected object and center on it (on pkgdb synteny object)
The highlight functionality has also been improved, a dark background will appear on the selected object . It is triggered by using “Center” option or “MoveTo“.
Please report us any bugs you may meet.
MicroScope: Genome browser improvement