University of Nantes – Master 2 Bioinformatics – 2015-2016
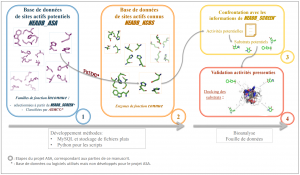
This report presents the work I carried out during my internship in research unit UMR8030 of the Genoscope institute. This unit aims at combining bioinformatics and high-throughput experimental approaches for the discovery of novel enzymatic activities in the metabolism of bacteria. It should be noted that nearly 35% of the proteins from large-scale sequencing of microbial genomes are annotated with unknown function. The objective of my internship was to explore, by analogy, active sites from proteins of unknown function using 3D tools in order to suggest enzymatic activities. This work was carried out in four stages. The first stage involved was the creation of a database containing potential active sites generated by predictive software that localizes the active sites of an enzyme family and classifies that family into sub-families. The second stage combined: (i) the building of a second database containing actives sites from enzymes with known function; and (ii) the comparison of these two databases using a 3D pattern tool search. The goal of the third stage was to validate the predictions made by the software by running docking simulations of potential substrates on representatives of protein families. Finally, the pipeline was validated on two families with known function and tested on nine families of unknown function. The protocol highlighted some new aldolases and mutarotases activities for two families. These enzymatic activities will soon be experimentally tested by a team from the UMR.
Keywords : Enzymatic activities, Proteins families, Database, Analogy of active site