!!! Thank you very much for your active participation to the last MicroScope survey !!!
Conducted in summer 2014 (till November), this survey intended to gather information about our users (occupation, frequency of use, most common use cases, etc.), their satisfaction about existing tools and services, and their needs (enhancement of existing functionalities, additional features/tools, speed of genome integration, and update/storage of data). Some questions were concerned with MicroScope users perceptions on (i) the position of MicroScope compare to its competitors, (ii) MicroScope current main originalities and necessary evolutions, and (iii) an invoicing of part of the MicroScope services.
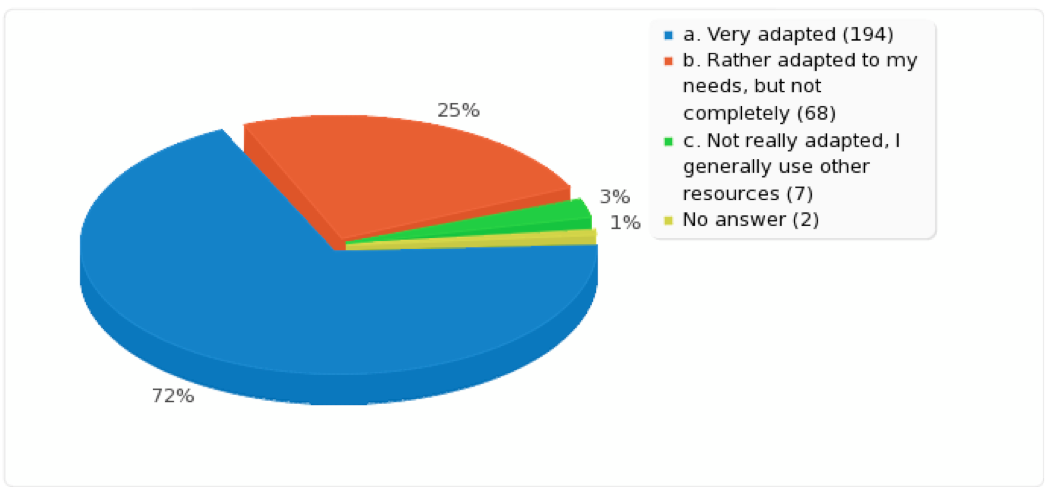
We very much appreciate all your input. We received 271 responses (on 2,298 actual user accounts of which about 600 are really “active” ones) containing many detailed comments and interesting suggestions. An overview of the quantitative results and a summary of your comments and suggestions from the free-text questions can be found here.
In addition, to answer important comments raised in this survey, this file addresses more specifically the following points:
- Why using the MicroScope platform (i.e., instead of other tools like the RAST server)?
- Several required functionalities are already implemented in the platform
- About “missing” bacterial genomes in PkGDB and synteny computations
- Actual slow process in integrating bacterial genomes and/or updating blast results
- What about the future MicroScope economic model?
If you would like further information about specific requests, please do not hesitate Contact Us (mage@genoscope.cns.fr).
Linked to several mandatory technological improvements of the MicroScope platform we currently prioritize the following developments/improvements:
- Increase the speed of genome integration
- Deployment of MicroScope in the Cloud: toward a Software as a Service for on-demand analyses of microbial genomes
- Data compression for Core/Pan genome representation and synteny computation
- Genome selection using taxonomic classification
- Propagate the annotation curation when working on multiple clones of the same species or closely related species
- Integration of metadata on Organisms like growth phenotypes
