 We are pleased to announce the release of a new tool dedicated to the computation of Pan/Core genomes. This development of this tool is an answer to requests we have had during the MicroScope satisfaction survey 2012.
We are pleased to announce the release of a new tool dedicated to the computation of Pan/Core genomes. This development of this tool is an answer to requests we have had during the MicroScope satisfaction survey 2012.
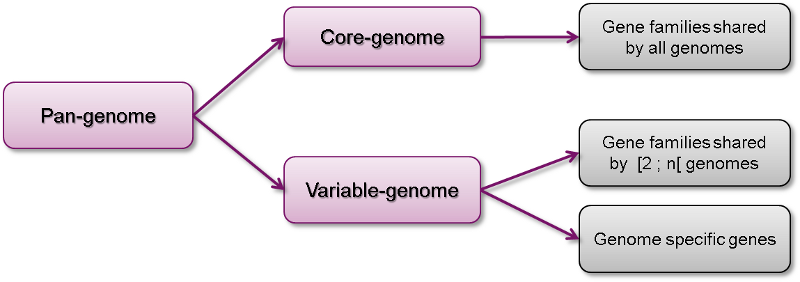
The Pan/Core interface provides an analysis of the pan-genome and its components (core-genome, variable-genome) for a set of organisms. It uses the MicroScope gene families (MICFAM) which are computed with the SiLiX software (« Ultra-fast sequence clustering from similarity networks with SiLiX. », Miele V et al., 2011).
It allows the users to:
- Compute pan-genome and core-genome sizes and their evolutions, for a set of selected genomes (up to 200)
- Determine the proportion of common and variable genome for each genome
- Extract core-genome, variable-genome and strain specific sequences and annotations.
http://www.genoscope.cns.fr/agc/microscope/compgenomics/pancoreTool.php
Your feedbacks are welcome!
Reference: Miele V, Penel S, Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011 Apr 22;12:116.