The carbohydrate-active enzyme (CAZyme) prediction tool has been updated for dbCAN3 & run_dbcan 5.2.1 on the MicroScope platform.

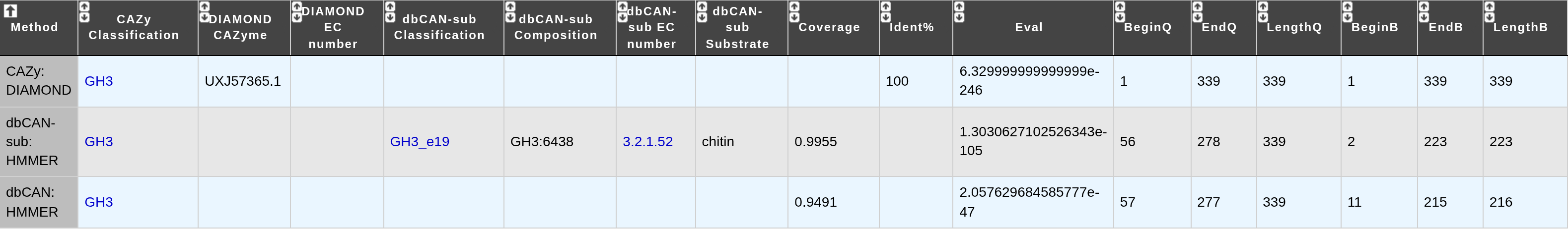
The tool continues to use DIAMOND (for sequence comparison against the CAZy database) and HMM (for comparison against CAZy families HMM profiles) but Hotpep has been replaced by dbCAN-sub. dbCAN-sub is a profile Hidden Markov Model database (HMMdb) for substrate prediction at the CAZyme subfamily level. Each dbCAN-sub HMM corresponds to an eCAMI subfamily, containing protein sequences annotated by CAZy, some of which may have been experimentally characterized with EC numbers. This enables substrate prediction and provide subfamily-level annotations enriched with EC numbers and subfamily composition.
As a result, this new version offers clearer and more detailed output. These prediction results can be seen in Genomic Object Editor and queried in the Search by keyword functionality. You may also refer to the dbCAN section of the MicroScope User documentation for further information.
Jinfang Zheng , Qiwei Ge , Yuchen Yan, Xinpeng Zhang, Le Huang, Yanbin Yin, dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023 Jul. https://doi.org/10.1093/nar/gkad328
Please report us any bugs you may meet.
