Master 2 – Phylogenomic analysis of Topo6 in archaea
Description
Location:
You will be working in the Bioinformatics Analysis Laboratory for Genomics and Metabolism (LABGeM, UMR8030, https://labgem.genoscope.cns.fr) at Genoscope (Institut de biologie François Jacob – CEA – Evry), a scientific research centre dedicated to environmental genomics. The LABGeM is interested in analysing and exploring the diversity of microorganisms and their metabolism. Our team has expertise in the annotation and comparison of prokaryotic genomes through the development of several approaches and methods, particularly in genome-wide analysis (such as PPanGGOLiN1, panRGP and panModule2). It also offers an international community of microbiologists access to a web-based platform called MicroScope3.
Scientific context :
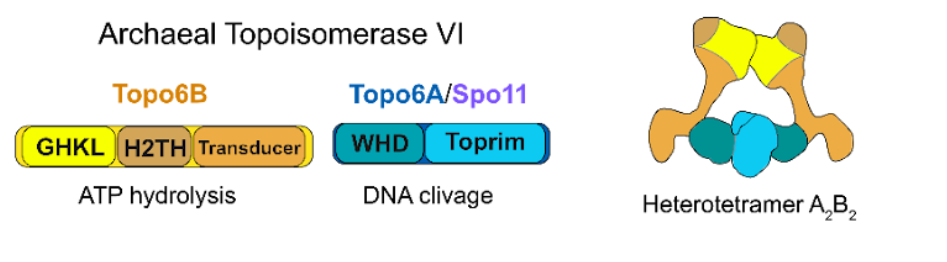
Eukaryotic SPO11 proteins catalyze the formation of meiotic DNA double strand breaks and are evolutionary related to subunit A of archaeal Topoisomerase VI (TopoVI)4,5. We have recently discovered that Asgard archaea, suggested as the closest known relatives of Eukaryotes6,7, encode SPO11-like proteins in addition to TopoVI. ARCHSPO11 aims to characterize these archaeal SPO11 proteins by using phylogenomics and in vitro biochemical assays. These studies should shed light on the emergence, evolution and molecular mechanism of meiotic recombination pathway, a key step in sexual reproduction. The objectives of the ARCHSPO11 are two fold: (i) assess the diversity and phylogeny of TopoVI subunits in available Asgard genomes; (ii) express and purify TOPOVIL-like complexes from Asgard archaea and determine their biochemical properties in vitro (will be done by the collaborators).
Objective :
The aim of the master project is to assess the diversity of TopoVI in Asgards genomes. The phylogenetic diversity of Asgard archaea was substantially expanded in the last couple of years 6,7 raising from a dozen to 326 genomes. The ARCHSPO11 will benefit from this wealth of novel genomic data to draw up an exhaustive classification of TopoVI subunits and determine their evolutionary trajectories.
Startegy :
- We will use pangenomic approach on Asgards genomes (i) to considerably reduce the bias of using a single representative genome per species or genus and (ii) to eliminate redundancy by removing strain specific genes.
- We will create profile hidden Markov models (HMM) of known Topo VI orthologs and match them against the Agards pangenomes database to generate ‘presence-absence’ phyletic patterns across the Asgard diversity.
- Potential Asgard Topo VI predicted from the homology searches, and HMM profile search, will be used to perform protein similarity network and phylogenetic analysis and, in fine, to discriminate SPO11-like proteins from the classical Topo VI. For SPO11-like encoding genes, we will use “guilt by association” strategy by analyzing genomic co-localization and synteny to search for functionally related proteins.
- Structural models for SPO11-like proteins versus typical Topo VI will be predicted using AlphaFold2 and ESMFold.
Profile required :
Master 2 in bioinformatics

- Programming (python, bash, R)
- Good knowledge of genomics
- Knowledge in microbiology and phylogenomics
Contacts :
Violette Da Cunha (vdacunha@genoscope.cns.fr)
Financial Support :
Living Machines @ Work
References
1. Gautreau, G. et al. PPanGGOLiN: Depicting microbial diversity via a partitioned pangenome graph. PLoS Comput Biol 16, 1–27 (2020).
2. Bazin, A., Medigue, C., Vallenet, D. & Calteau, A. panModule: detecting conserved modules in the variable regions of a pangenome graph. bioRxiv 2021.12.06.471380 (2021).
3. Vallenet, D. et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res 48, D579–D589 (2020).
4. Vrielynck, N. et al. A DNA topoisomerase VI–like complex initiates meiotic recombination. Science 351, 939–943 (2016).
5. Robert, T. et al. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351, 943–949 (2016).
6. Liu, Y. et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 1–5 (2021) doi:10.1038/s41586-021-03494-3.10.
7. Xie, R. et al. Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes. Sci. China Life Sci.65, 818–829 (2022).
